Science Spyglass
Next generation
neuroscience drug screening
Tessara's RealBrain® neural micro-tissues validate compound effects in Axxam’s High-Content Imaging workflow
Application note
Axxam S.p.A. – Milan (Italy)
Tessara Therapeutics Pty Ltd. – Melbourne (Australia)
Keywords: chemotherapy induced neuropathy (CIPN), induced pluripotent stem cell (iPSC), neurotoxicity, high content imaging, mini brain, organoids, drug discovery, neural micro-tissues, neurodegenerative diseases.
Introduction
The use of three-dimensional (3D) biology has revolutionized the way to model and study complex biological processes, surpassing the traditional two-dimensional (2D) approaches. While 2D cell cultures have been the standard for many years, they fail to capture the intricate cell-to-cell and cell-to-matrix interactions that occur in vivo.
In contrast, 3D biology offers a more physiologically relevant environment, allowing for the study of cellular behaviors in a context that mimics in vivo conditions more closely. The growing need for non-animal models in biomedical research is also driving the shift toward 3D systems. Indeed, in 2022, the U.S. FDA made headlines by endorsing non-animal testing methods for drug discovery and development, emphasizing the importance of human-relevant models for in vitro drug discovery.
Whilst the global life science community has begun rapidly expanding and adopting 3D human tissue models such as spheroids and organoids, a number of limitations have arisen. Spheroids for example are reproducible, but like 2D cultures, the structure of their neural networks is markedly different from the human brain. Organoids can mimic brain complexity but have lacked reproducibility and have been difficult and slow to manufacture. In response to this demand and to facilitate the uptake of 3D models that can better meet the requirements of a preclinical drug discovery pipeline, a new technology from Tessara Therapeutics was introduced.
Tessara's 3D models combined with Axxam’s workflow
Based in Melbourne Australia, Tessara is a world leader in 3D cell-based models of the brain. In a world-first innovation, Tessara has developed its RealBrain® neural micro-tissue technology and 3D cell-based models that replicate the biological complexity of the human brain in a reproducible manner. Tessara has validated and recently launched two brain models: the ArtiBrain™ model (healthy human brain), and the ADBrain™ model (sporadic Alzheimer’s disease (AD), representing ~98% of all AD cases). Tessara has developed this platform with commercial scale manufacturing processes, delivering models that are predictive, reproducible at scale and cost-effective.
Combining these relevant in vitro models with suitable approaches to fully harness their potential could be transformative for the drug discovery field. In this context, high-content screening (HCS) and image-based readouts represent ideal tools for exploring the dynamic behaviours of cells and networks within 3D environments. Through image-based analysis, researchers can explore the complex biology of 3D systems in unprecedented detail across multiplecellular parameters, generating vast datasets from complex biological systems, facilitating the identification of subtle phenotypic changes that would be missed in traditional screening methods. This technology pipeline enables a deeper understanding of drug responses, toxicity, and mechanisms of action in more relevant biological contexts, offering greater translational value for pre-clinical studies. Incorporating these advanced technologies into research pipelines is transforming the drug discovery and disease modeling field.
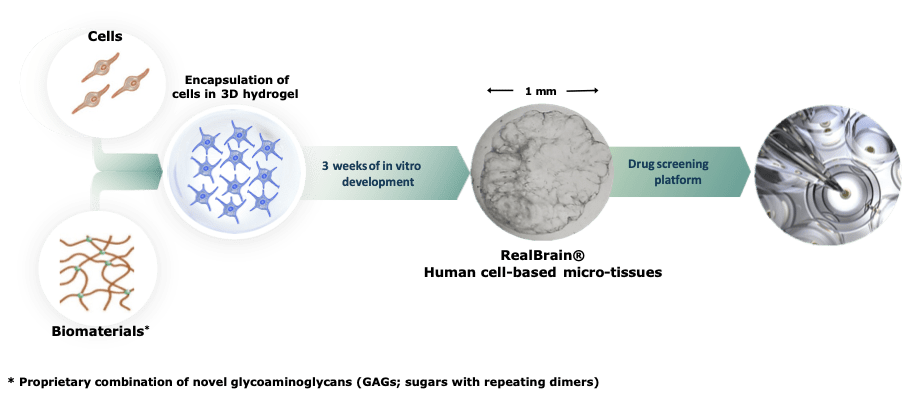
Generation of Tessara’s RealBrain® neural micro-tissues
Tessara’s RealBrain® drug screening platform is based on manufactured human mimetic brain micro-tissues that closely recapitulate neural physiology and pathophysiology. RealBrain® neural micro-tissues are generated by encapsulating primary or induced pluripotent stem cell (iPSC)-derived neural precursor cells in a proprietary hydrogel matrix composed of chemically defined biomaterials. The RealBrain® hydrogel provides the optimal micro-environment to activate endogenous cellular programs of neuro-development in vitro. Over three weeks of maturation, the developing neural and glial cells remodel their micro-environment, migrate in 3 dimensions, form a complex network and replace the synthetic hydrogel with their own cell-secreted extracellular matrix (ECM). The resulting RealBrain® neural micro-tissues closely model human neurophysiology – they respond to neurotransmitters, feature a genetic profile representative of human brain tissue, and under controlled conditions can develop the complex pathological hallmarks of Alzheimer’s disease. These neural micro-tissues are also compatible with all standard laboratory workflows, from immunocytochemistry and confocal microscopy that doesn’t require the use of any clearance protocols, through to genomics and proteomics applications and other functional assays such as calcium imaging.
Imaging the RealBrain® micro-tissues with Axxam's High Content Imaging workflow
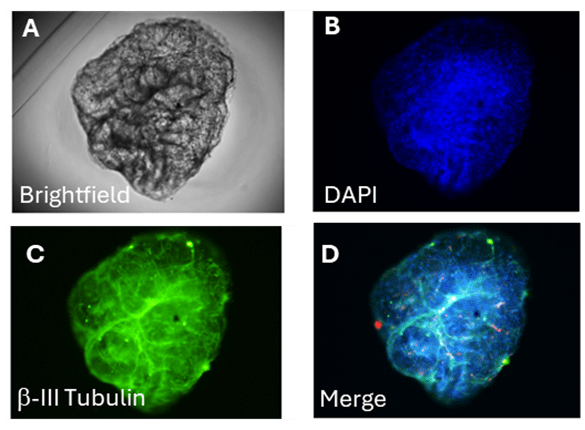
In this study, we employed an automated high-content confocal imaging system (Opera Phenix Plus, Revvity) to acquire detailed 3D images of RealBrain® micro-tissues, stained with fluorescent markers to visualize neuronal cells (β-III Tubulin), glial cells (GFAP) and cell nuclei (DAPI) (Figure 1); these images illustrate the structural organization and neuronal differentiation within the brain micro-tissue.
Figure 1
Representative images of a brain micro-tissue acquired by High Content Microscopy (Opera Phenix Plus, Revvity). (A) Brightfield image showing the overall morphology of the microo-tissue. (B) DAPI staining (blue) highlighting cell nuclei distribution. (C) βIII-Tubulin staining (green) marking neuronal structures. (D) Merged image combining DAPI and βIII-Tubulin signals, providing a composite view of the organoid’s neuronal architecture.
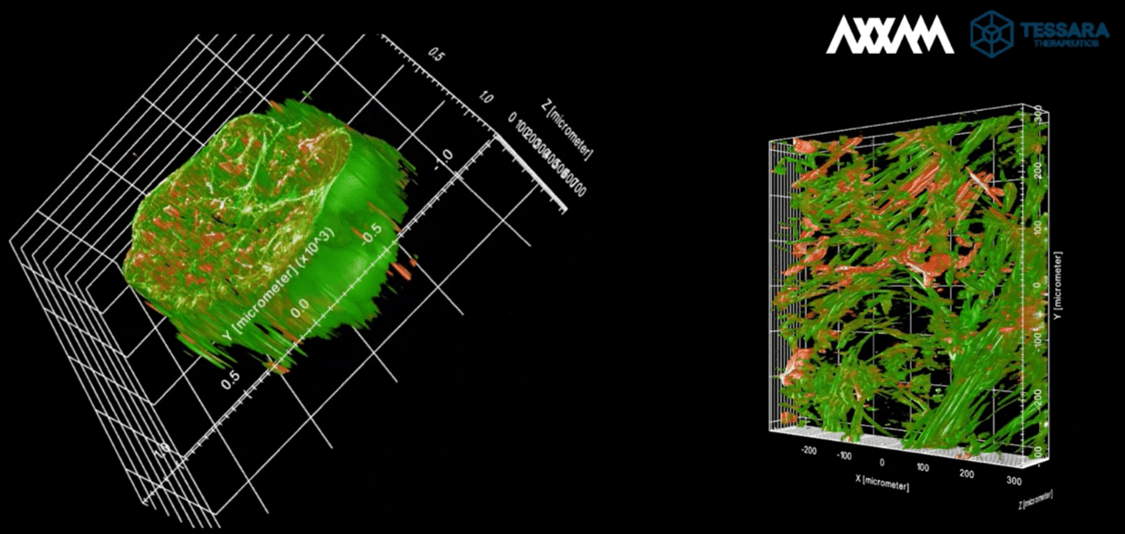
Using two different acquisition setups and z-stack acquisitions we were able to capture multiple optical slices, enabling comprehensive 3D reconstruction of the neural micro-tissues at different levels of resolution (Figure 2).
Figure 2
High-resolution 3D imaging of microtissues acquired through z-stack acquisition, enabling detailed visualization of cellular structures at different depths. Left panel: A volumetric reconstruction of the microtissue, showcasing its overall morphology and spatial distribution of cellular components. Right panel: Cross-sectional view highlighting internal structural organization, revealing intricate cellular networks and fiber orientations. Neuronal cells are stained by βIII-Tubulin (green) whilst glial cells are marked by GFAP staining (red).
Key parameters such as z-stack step size, magnification, field of view, and laser intensities were carefully optimized to ensure both signal clarity and high-quality micro-tissue reconstruction. We balanced acquisition time and resolution to maintain both data quality and efficiency. The resulting images were analyzed using Harmony software (Revvity), where we developed a customized pipeline to visualize cellular features and extract multiparametric data. To assess the impact of magnification on data quality, we compared two settings: 20X (high magnification) and 5X (low magnification) (Figure 3).
Figure 3
Examples of acquisition parameters (right panel) and representative images of the field/well coverage depending on the magnification applied. Top right (5X magnification): Whole micro-tissue overview, providing structural context and overall morphology. Bottom right (20X magnification): High-resolution imaging revealing detailed cellular organization and fiber structures, enabling more precise analysis of cellular components.
By using 5X magnification, the entire micro-tissue could be captured within a single field of view (FoV), requiring around 100 Z-stacks (spaced 8 µm apart) to cover the full volume across 4 channels (Brightfield, Hoechst, A488, and A568). This strategy reduced the acquisition time to 0.5 hours with a total data size of 10GB for the 9 samples tested, compared to 20X acquisition. This low-magnification setting enables a fast acquisition, and it is amenable for high-throughput analysis.
By using 20X magnification, we used a total of 10 Z-stacks spaced 8 µm apart to capture the central structure of each micro-tissue. 49 FoVs were imaged per sample across 4 channels (Brightfield, Hoechst, A488, and A568). These 49 FoVs were stitched together to reconstruct the entire micro-tissue. The acquisition time was doubled compared to 5X acquisition setting with a data size of 44GB for the 9 samples tested. This approach provided a good balance between high resolution and time efficiency while keeping data manageable for processing and storage. This flexible acquisition strategy allowed us to adapt our imaging approach according to the desired balance between resolution, acquisition time, and data management.
A key advantage of this methodology was the ability to reconstruct the entire 3D structure of neural micro-tissues and, together with its superior transparency, we were able to provide a more detailed and accurate analysis of cellular behaviors and interactions within these complex models.
Image analysis was subsequently performed on the z-stacks maximum projection and carried out in Harmony software (Revvity). We developed an ad-hoc pipeline to explore several features for both the low and high magnification acquisitions. Key features extracted from image analysis were categorized into nuclei detection and image region properties such as size, shape, and fluorescence intensity. Metrics included area, perimeter, width, length, and roundness, providing structural insights. Fluorescence-based features capture signal intensity and distribution, highlighting edges, ridges, and bright regions of the micro-tissues.
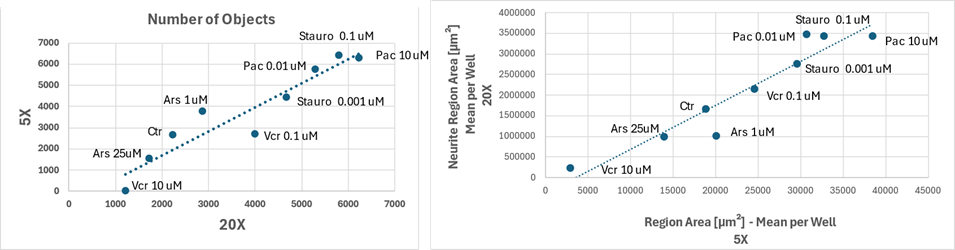
Figure 4 Features comparison, object count (left) and neurite region area (right), across 5X and 20X magnifications for different drug treatments and concentrations
Image analysis results remained consistent across magnifications, supporting the reliability of feature quantification despite differences in resolution, as shown in Figure 4 where image features compared between drugs and concentrations at the two magnifications: each data point represents a specific condition, and the dotted trendlines indicate a strong correlation between the two magnifications. In both graphs, the distribution of points along the trendline suggests that the numerical values of the features extracted from the two imaging settings are well aligned.
Use of Tessara's RealBrain® micro-tissues to visualize chemotherapy induced neuropathy (CIPN) and neurodegeneration using imaging at Axxam
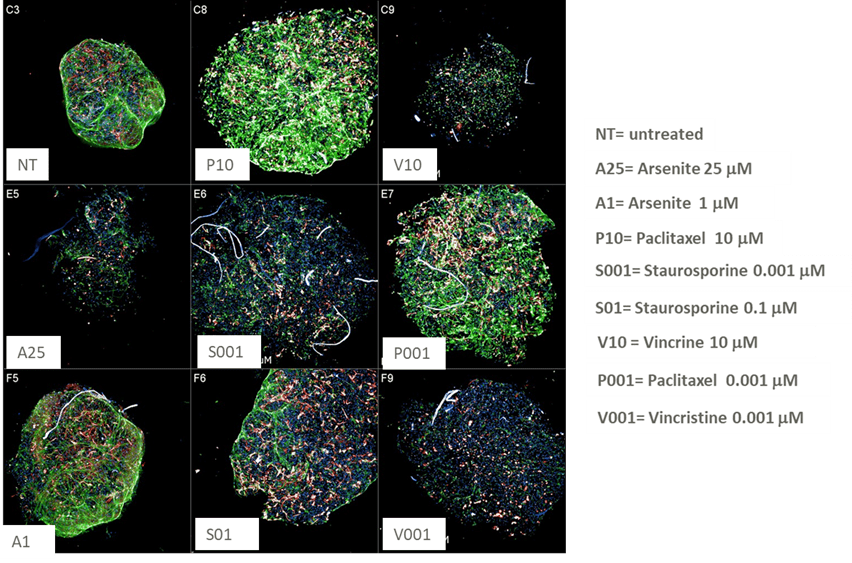
We can use the model to capture changes in morphology/markers expression following compound treatments that can mimic several neurotoxicity conditions. We selected four compounds known for their toxicology (arsenite, paclitaxel, staurosporin, and vincristine) and we tested them on the micro-tissues using 3 different concentrations. Figure 5 shows a representative dose-response of paclitaxel and the effect of this drug on different markers in the specific channels (Brightfield, DAPI, A488-βIII-Tubulin, and A568-GFAP). Increasing the concentration of this drug leads to a significant disruption of the neuronal network structure, accompanied by noticeable enlargement and loss of integrity of the microtissue surface. Similar neurotoxic effects are observed with other tested compounds, aligning with previously reported data in literature. These findings underscore the suitability of 3D culture models in capturing complex structural and network-level phenotypes in neurotoxicity studies.

Figure 5 Representative images of neural micro-tissues treated with increasing concentrations of Paclitaxel.
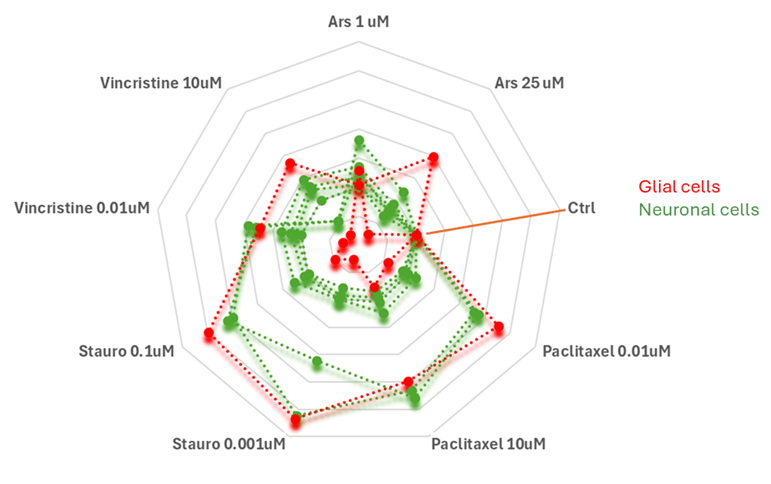
To analyze the impact of the different drug treatments on the features of the neural micro-tissues, we performed a radial analysis in order to compare multivariate data (aggregate features) obtained from different drug treatments in the co-culture system of glial cells (red) and neuronal cells (green). Specifically, each axis of Figure 6 represents a specific drug condition, with varying concentrations as indicated. The control condition (Ctrl) shows the closest clustering of both cell types, suggesting a baseline similarity in their aggregate features. Upon treatment, the profiles of neuronal and glial cells diverge, highlighting distinct cellular responses. In particular, Staurosporine (Stauro) and Vincristine treatments appear to induce more pronounced changes in glial cells, as indicated by their extended red profiles. Paclitaxel and arsenic (Ars) treatments show differential effects, where neuronal cells exhibit stronger deviations, particularly at certain concentrations.
Figure 6
Radar plot comparing aggregate features of glial cells (red) and neuronal cells (green) under different drug treatments. Each axis represents a specific drug condition, with different concentrations. The control condition (Ctrl) serves as a baseline. Divergences in the profiles indicate differential cellular responses to treatment.
Investigating the single morphological features, Area and Perimeter of the micro-tissue, stand out with higher relevance for these drugs compared to control wells. The increase in the area and perimeter can be attributed to the loss of structure and swelling of the micro-tissue in response to the toxicity of the drug, as can be seen clearly in Figure 7.
Future directions and opportunities
This collaboration has demonstrated the utility of the RealBrain® neural micro-tissue platform and its application to preclinical drug screening pipeline. With the ability to develop bespoke models (of not only different disease indications, but also the healthy brain at different developmental stages) in combination with the capacity for scalable and robust high-content analysis and other applications, then this presents a versatile platform for screening small molecules and other biologics in the context of human disease. Such deep insights in a human-relevant model will be a catalyst for continued development and adoption of non-animal models as the industry moves towards the utilization of platforms and models that are better able to predict drug efficacy in humans and thus, have greater clinical translation.
- Wheeler HE et al. Modeling chemotherapeutic neurotoxicity with human induced pluripotent stem cell-derived neuronal cells. PLoS One. 2015. PMID: 25689802.
- Schinke C, et al. Modeling chemotherapy induced neurotoxicity with human induced pluripotent stem cell (iPSC) -derived sensory neurons. Neurobiol Dis. 2021.PMID: 33984509.
Application note in collaboration with

Tessara Therapeutics Pty Ltd.
Jumar Bioincubator
Level 11, 655 Elizabeth St,
Melbourne, Victoria, 3000 – Australia
Tessara’s contacts:
Professor Paul Adlard
Chief Scientific Officer
paul.a@tessaratx.com
Tessara’s contacts:
Dr Christos Papadimitriou
Chief Executive Officer & Managing Director
christos.p@tessaratx.com